Section: New Results
Medical Image Analysis
Brain tumor cell density estimation from multi-modal MR images based on a synthetic tumor growth model
Participants : Ezequiel Geremia [Correspondant, Inria] , Nicholas Ayache [Inria] , Antonio Criminisi [MSRC] , Bjoern Menze [Inria,ETHZ] , Marcel Prastawa [University of Utah] .
Published in the proceedings of the MCV Workshop at MICCAI 2012 [36]
biophysiological tumor growth simulator, multi-variate regression random forests, gliomas, MRI
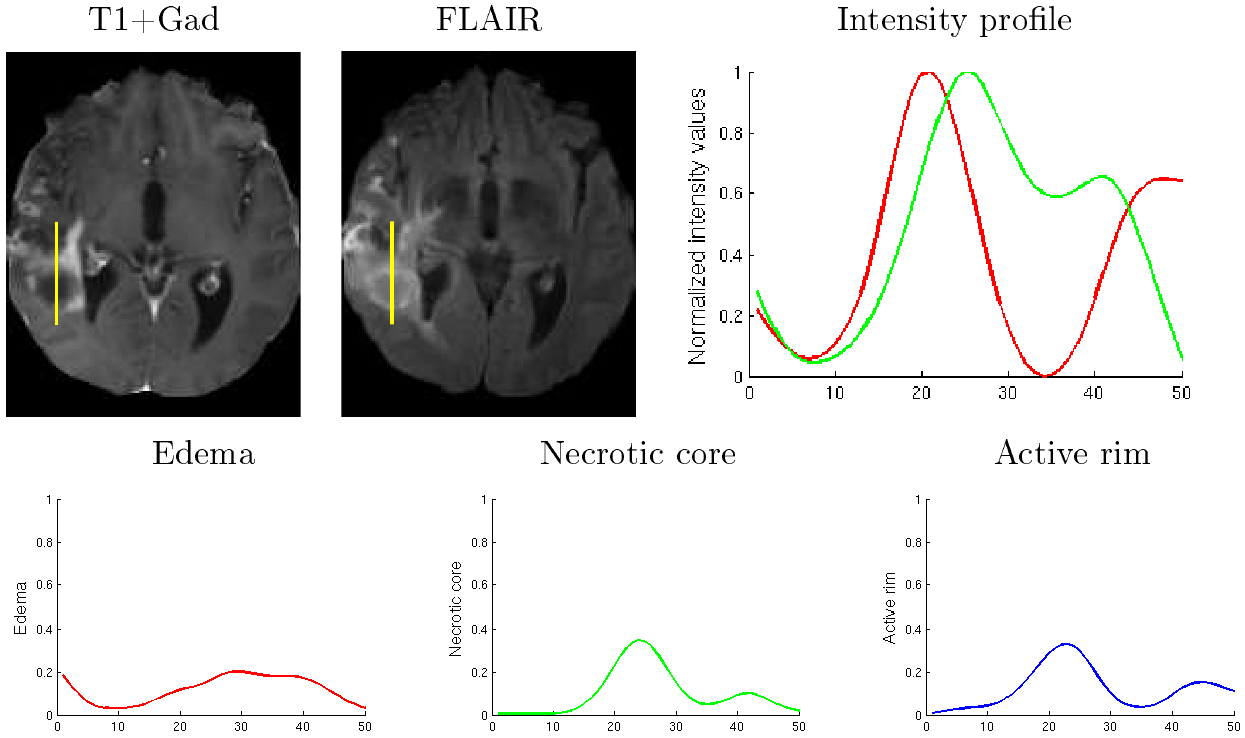
A generative-discriminative framework is presented to learn model-based estimations of the tumor cell density
The ground truth for tumor cell density is very hard to obtain
A biophysiological tumor growth simulator is used to generate the ground truth tumor cell densities and associated MRIs
A multi-variate regression random forests is trained to estimate the voxel-wise distribution of tumor cell density from input MR images
The training data contains 500 synthetic cases and their associated ground truth generated by the brain tumor simulator
The method was tested on 200 synthetic cases with excellent results
The method also provided very promising results for estimating the tumor cell density on 16 clinical cases showing low grade gliomas from the DKFZ (German Cancer Research Center)
|
Automatic indexation of cardiac MR images
Participants : Jan Margeta [Correspondant] , Nicholas Ayache, Antonio Criminisi [MSRC] .
This work has been partly supported by Microsoft Research through its PhD Scholarship Programme and the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Machine learning, Cardiac MR, MR preprocessing
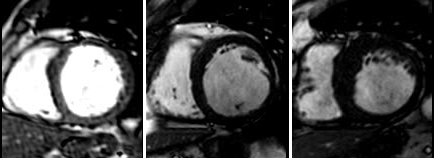
A generic random forest framework has been implemented and its recent modifications have been applied to a fully automatic and a semisupervised image segmentation methods, and manifold learning in cardiac MRI.
We have performed image based cardiac function quantification from preprocessed cardiac cine MRI sequences.
An image intensity standardization in magnetic resonance images method has been proposed.
Multimodal brain tumor segmentation
Participant : Bjoern Menze [Correspondant] .
MICCAI 2012
Further developed the generative brain tumor segmentation model
Developed a generative-discriminative model for multimodal brain tumor segmentation
Developed a new regularization approach for longitudinal tumor segmentation (with Guillaume Charpiat & Yuliya Tarabalka, Inria Sophia-Antipolis)
Initiated and co-organized an international benchmark on multimodal brain tumor segmentation as a challenge workshop during MICCAI 2012 in Nice (http://www2.imm.dtu.dk/projects/BRATS2012 )
Statistical Analysis of Diffusion Tensor Images of Brain
Participants : Vikash Gupta [Correspondant] , Xavier Pennec, Nicholas Ayache.
Diffusion Tensor Imaging of Brain, Tractography, Super-resolution, Statistical analysis
Diffusion tensor imaging (DTI) is gaining interest as a clinical tool for studying a number of brain diseases pertaining to white matter tracts and also as an aid in neuro-surgical planning. Unfortunately, in a clinical environment, the diffusion imaging is hampered by the long acquisiton times, low signal to noise ratio and a prominent partial volume effect due to thick slices. The present work aims at robustifying the analysis of clinical images by developing a super-resolution algorithm for DTI and quantifying its improvements with respect to the existing tensor estimation methods. Part of the work was presented at the 1st International Symposium on Deep Brain Connectomics [69] .
3D/2D coronary arteries registration
Participants : Thomas Benseghir [Correspondant] , Grégoire Malandain, Régis Vaillant [GE-Healthcare] , Nicholas Ayache.
This work is done in collaboration with GE-Healthcare (Buc).
3D/2D registration ; coronary arteries ; Chronic Total Occlusion ; X-ray fluoroscopy / CT image fusion
The context of this work is to provide the cardiologist with an advanced guidance application, where a pre-operative 3D CT segmented image will be superimposed on the per-operative 2D live fluoroscopy. Since the relative positions of the 3D image and the 2D projective images are unknown, we are currently investigating robust pose estimation methods before using an upcoming registration algorithm.